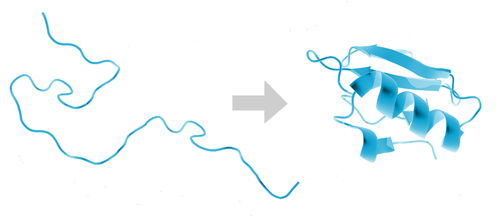
Our non-critical server systems are now running protein folding simulations in their spare time, as part of Stanford University’s Folding@home project. Understanding how proteins fold has important applications in medicine and molecular biology.
The project’s distributed computing cluster is currently one of the world’s most powerful at 21 petaFLOPS - an order of magnitude more than the world’s fastest supercomputer - and the results have contributed to over 100 scientific papers.
We have currently allocated 86 CPU cores across 5 nodes to the project, and may extend this to include underutilized production systems if it proves stable enough. You can follow our progress on our team page.
More from the blog
- Bengler Wins Jacob’s Prize 2016
- Simen made an opera about transhumanism*
- Deep Learning at the Museum
- Status / Arrivals
- Lego vs. Lego
- Spare Time Protein Origami
- GRBL with permissive license (MIT)
- We're building a 3d printer!
- Terrafab, now available in huge and tiny
- Checkpoint: Repcol
- Launch: Mapfest!
- Being in Nothingness
- Planning the brewery
